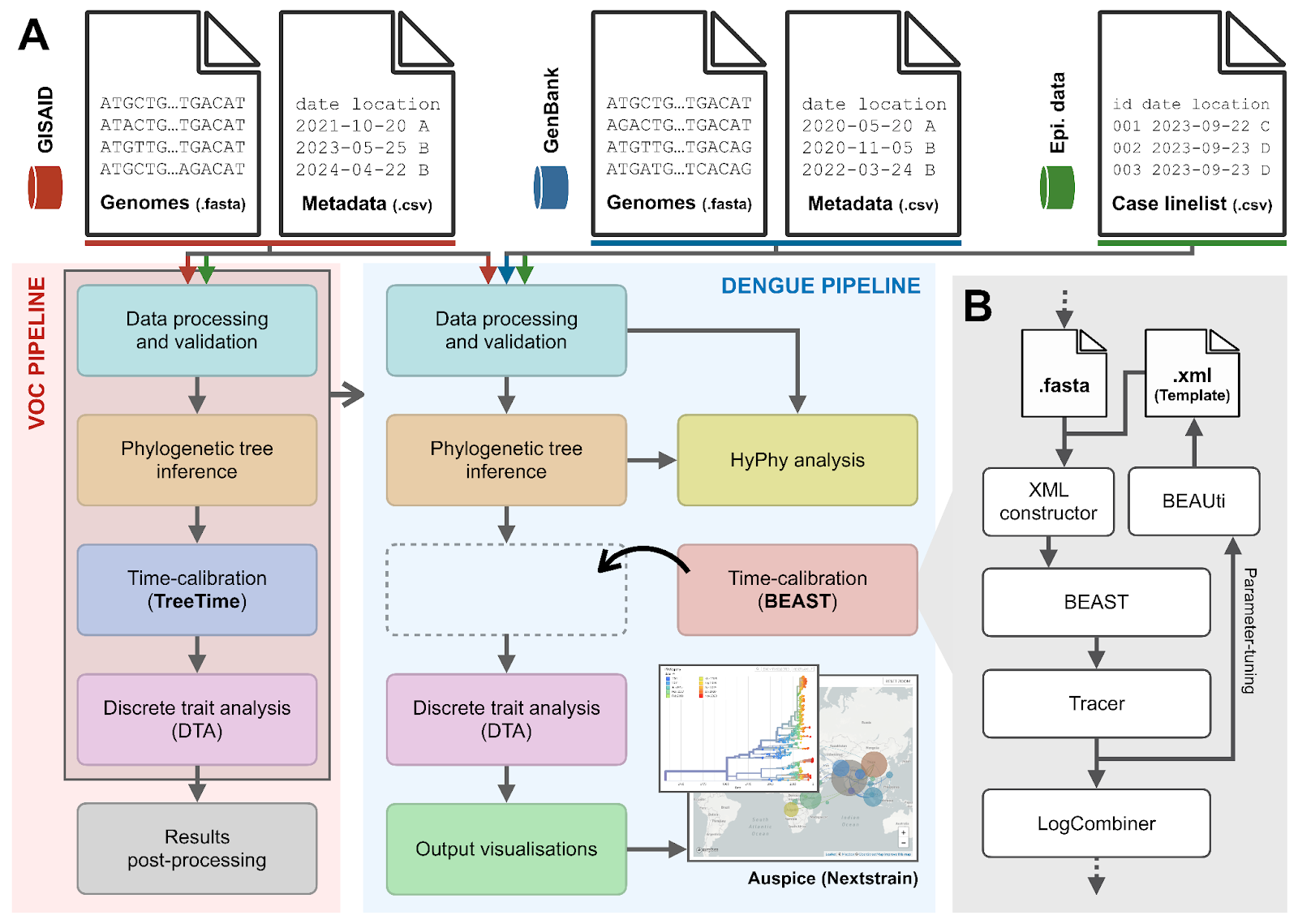
Graphical Analytical Pipeline Development Environment (GRAPEVNE) is an open-source, graphical pipeline development platform designed to streamline the construction, execution, and sharing of complex workflows in infectious disease research. GRAPEVNE simplifies interdisciplinary collaborations by providing an intuitive, modular interface for integrating epidemiological, genomic, and spatial data. By facilitating real-time outbreak analytics and pandemic preparedness, GRAPEVNE aligns with the global research community’s efforts to enhance data-driven public health decision-making.

GRAPEVNE – Graphical Analytical Pipeline Development Environment for Infectious Diseases
Currently in development, launching early 2021.