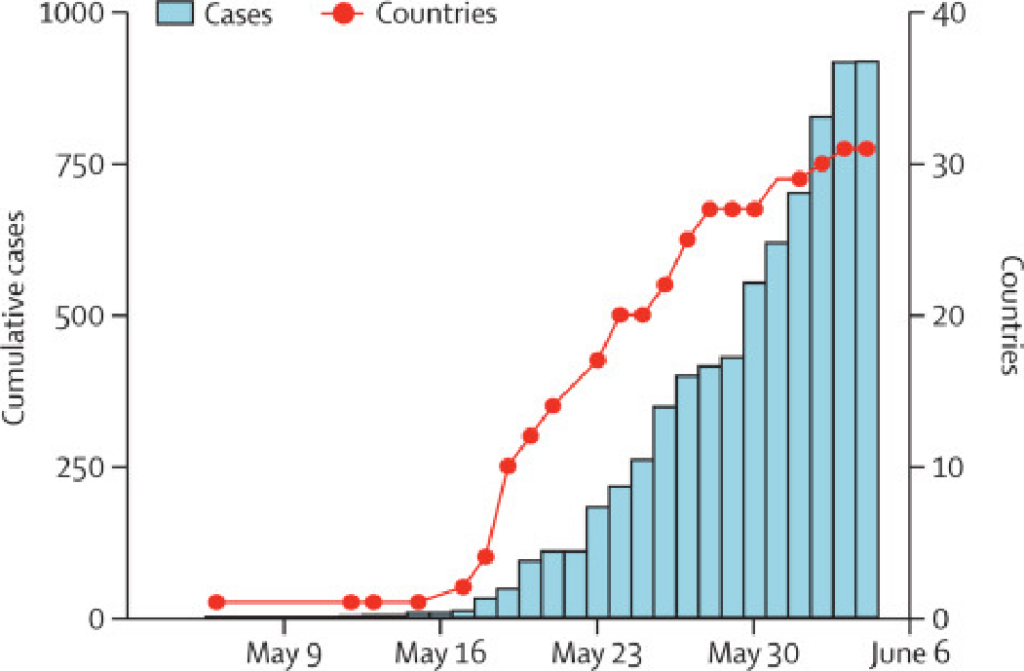
Monkeypox virus was first documented in humans in the 1970s and outbreaks have been reported in many countries, with most cases restricted to endemic areas.1 In early May, 2022, monkeypox cases were reported in the UK, Spain, and elsewhere in Europe (figure, appendix).2 The pattern of geographical dispersal was much larger compared with past outbreaks that were more localised and occurred often in under-resourced communities.3 The size of the outbreak clusters is growing each day, as is the geographical spread across Europe and North America. Within the first week of the initial report, 24 countries reported suspected and confirmed cases of monkeypox virus, some of which had known travel links to the UK, Spain, Canada, and western Europe. As of June 5, 2022, there have been 920 confirmed and 70 suspected cases. Of 64 confirmed cases with known travel history, 32 were associated with travel from Europe, three from west Africa, two from Canada, and one from Australia. For 26 cases, travel history locations remain unknown.
WHO convened a meeting of experts and technical advisory groups on May 20, 2022,4 to investigate the causes of the outbreak and have released updated guidance on surveillance, case investigation, and contact tracing.5 The reason for the outbreak having a broader geographical reach is being investigated by the international and national public health community and the research community, contributing to a finer scale understanding of the outbreak dynamics. However, cessation of smallpox vaccination programmes, encroachment of humans into forested areas, and growing international mobility seem to be playing important roles in the epidemiology of monkeypox virus outbreaks.6
To support global response efforts, our team created an open-access database and visualisation to track the occurrence of cases in different countries. In addition, where available, we added information on age (aggregated into age ranges, with a minimum range of 5 years), gender, dates of symptom onset and laboratory confirmation, symptoms, locations (aggregated to the state level), travel history, and additional metadata defined by WHO.5
Data are compiled from verified sources, including reports from governments and public health organisations and news media reporting of health official statements. As verified information and official statements are published, we document secondary sources and update the metadata in the dataset. An on-call schedule for curators that runs 24 h a day, 7 days a week was established to ensure data are updated in near real-time. Each case is seen and discussed by at least two curators before being made available via our Global.health GitHub repository, and pushed to the map visualisation at least four times per day.
During the early stages of outbreaks, obtaining reliable, synthesised data on the characteristics of cases is a challenge, especially at a global scale. Our work attempts to harmonise information across countries and provide additional data to support the epidemiological understanding of the origins and transmission dynamics of this outbreak. Ideally, these data are paired with virus genomic data and integrated directly with countries’ epidemiological line-list data. In our repository, we are also working with colleagues and the WHO Hub for Pandemic and Epidemic Intelligence to define a contact data schema allowing countries and researchers to estimate and re-estimate key epidemiological parameters, such as the incubation period and serial interval, across different settings.
Real-time data are necessary to plan effective control measures should this outbreak grow further. The work builds on infrastructure developed for epidemic control and pandemic preparedness and was used for the COVID-19 pandemic.7 Global efforts are needed to ensure similar efforts to rapidly harmonise and publish detailed epidemiological data are supported during future outbreaks of emerging and re-emerging pathogens. This example will be a learning pathway to build better surveillance systems globally.
MUGK and JSB report funding from The Rockefeller Foundation and Google.org. TdO received fees from Illumina as a member of the Infectious Disease Testing Advisory Board and received partial travel support to attend the Nobel Symposium of Medicine in May, 2022. IIB received consulting fees from BlueDot and NHL Players’ Association. All other authors declare no competing interests.